What is happening now...
After many years of studying polymers (dendrimers) for drug delivery and diagnosis, the lab has pivoted to explore the properties of ring-shaped molecules.
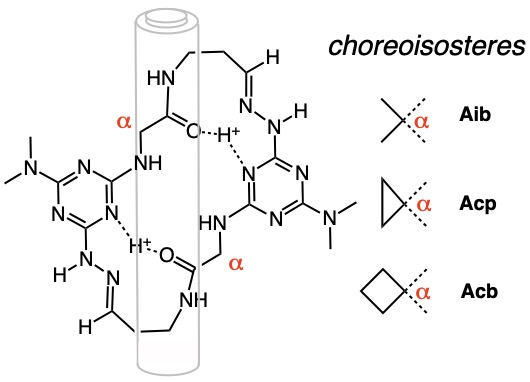
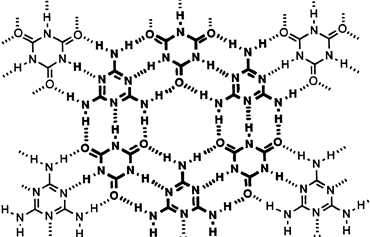
Nature provides effective medicines like cyclosporin and the erythromycins wherein the atoms are arranged in large rings. Making rings of atoms is not easy, but we have discovered a way to do so. Our current efforts focus on understanding the shapes that these molecules adopt and how they might be employed as drugs to fight diseases like breast cancer and Alzheimer's disease.
In a few simple chemical reactions, a crescent-shaped molecule can be prepared. Treating that molecule with acid leads to spontaneous dimerization and the formation of a macrocycle in excellent yields.
These macrocycles fold like door hinges. This behavior may allow them to transit membranes or strengthen materials.
For a list of graphical abstracts, click HERE.
RECENT WORK FROM TCU (2010-present)
As our efforts with dendrimer chemistry closed, we started studying ring-shaped molecules (macrocycles) and their application to drug design and materials science.
The Later Years at A&M (1998-2010): Mice "Cured" of Human Prostate Cancer
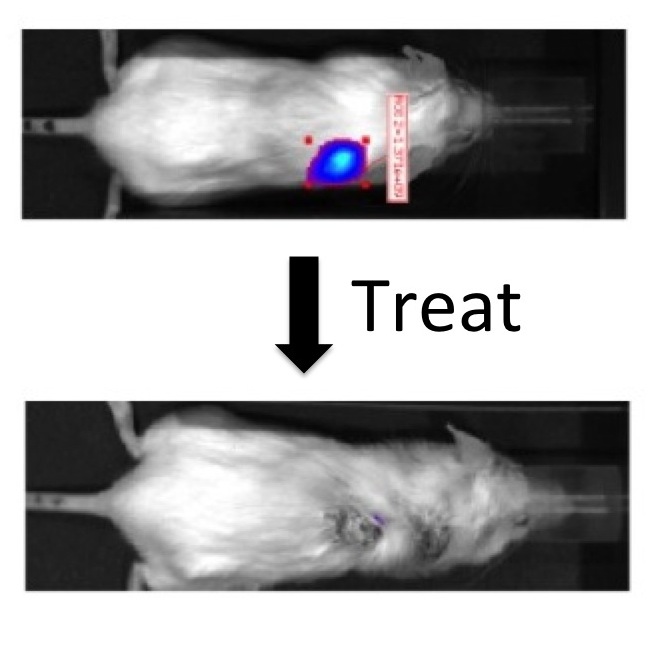
The capstone for our efforts was seeing a "cure" of prostate cancer in mice using a dendrimer that delivered the anti-cancer drug paclitaxel with collaborators JT Hsieh and Xiankai Sun at UT Southwestern Medical Center.
The Early Years at Texas A&M (1998-2010): Something Really Big
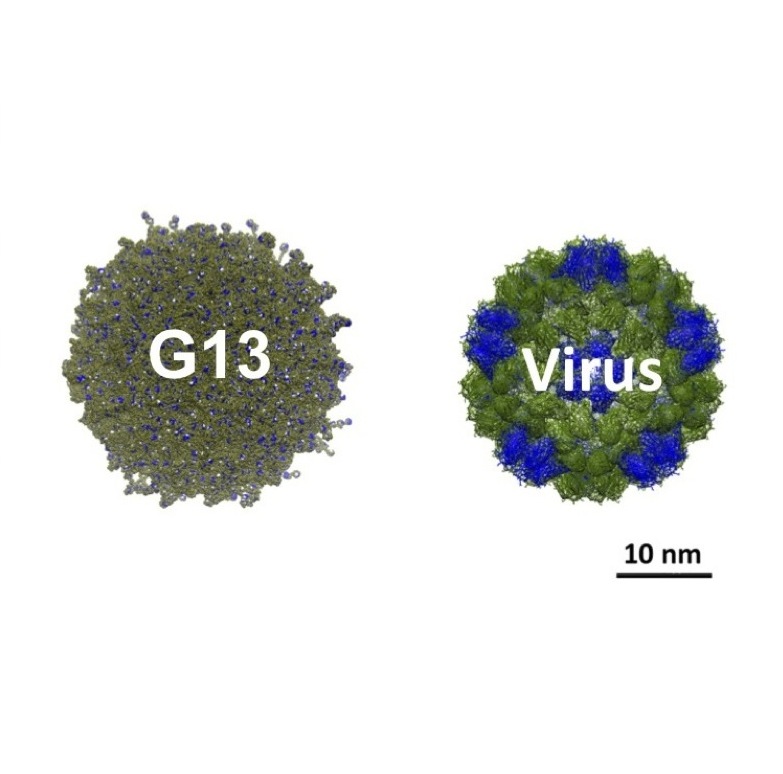
How big is big? When it comes to dendrimers, we have prepared one the size of a small virus: G13 has a theoretical molecular formula of C376,812H737,240N114,682O96,298 with a mass of 8.4 MDa.
POST-Doctoral work (1996-1998): A Sweet Turn
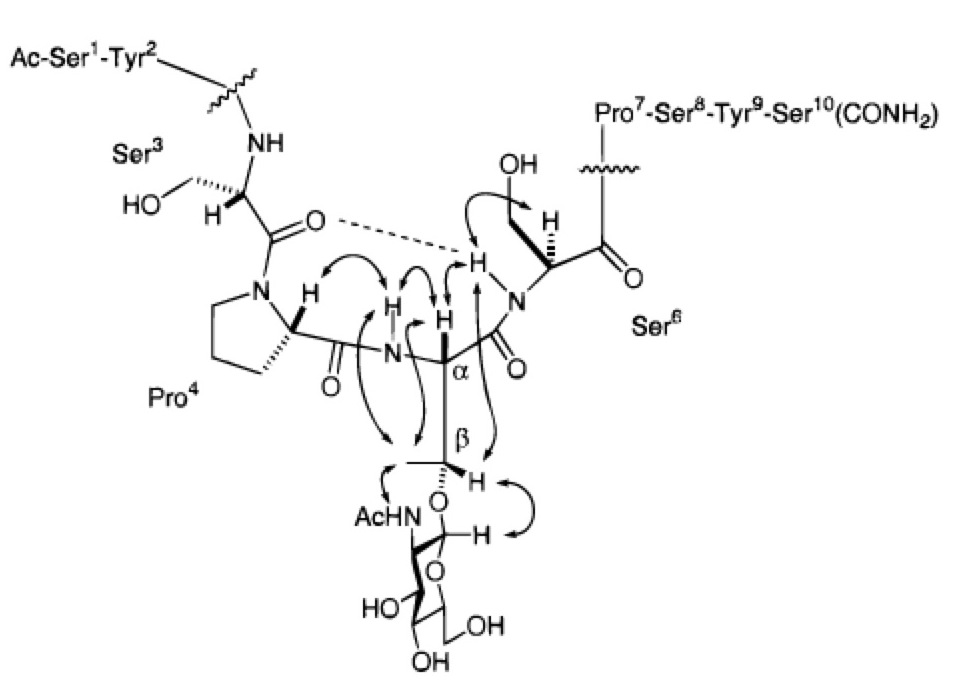
From post-doctoral studies, the conformation of one portion of the repeating tail of RNA Polymerase II forms a turn upon the addition of a single N-acetylglucosamine group.
GRADUATE WORK (1991-1996): Molecular Legos to Make Something Big
From graduate school, we held the record for the largest (most molecules), discrete supramolecular assembly for many years, a molecule we called Hub(M)9-9(CA) to capture the idea that 9 recognition domains (M, melamine) were tied into a single molecule through a central hub that recognized 9 other molecules (CA, cyanuric acid).
The longer story...
I entered the world of chemical research in 1989 as a sophomore at the University of Illinois in Urbana-Champaign by joining the laboratory of the late Professor Kenneth L. Rinehart, Jr. with the goal of contributing to their efforts of finding new drugs from the sea, so-called bioactive marine natural products. Mentored by Drs. George Robert Wilson, Ryuichi Sakai, John Carney and Chintamani Bapat, I worked to implement an L1210 cell toxicity assay and subsequently tried my hands at synthesis.
The experience opened the doors for summer experiences doing molecular biology at the USDA with the goal of isolating the gene for tricodiene synthetase in Tom Hohn's lab. Subsequently, I turned to synthesis in the anti-inflammatory group of Abbott Laboratories under Drs. Dan Kerkman and Joe Delaria.
After graduation, I pursued a PhD at Harvard University under the direction of Dr. George Whitesides wherein I was charged to "make something big" from smaller building blocks. The goal was to understand how molecules could be used as Legos to understand the balance between the stability of the thing that was built and the number of molecules required to build it. The field bears many monikers including molecular recognition, self-assembly, supramolecular chemistry, and crystal engineering whose meanings convey more subtle distinctions of the activities afoot. The camaraderie made the experience all the more amazing, and given the size of George's group, I'll abbreviate the acknowledgement to Drs. Chris Seto, John Mathias, John Folkers, Jim Wilbur, Milan Mrksich, Rich Wing, Andrew Black and Mathai Mammen. The journey was made richer by a great number of classmates; contemporaries, junior and senior.
From there, I joined the laboratory of Dr. Chi-Huey Wong as a post-doctoral fellow at The Scripps Research Institute where I was introduced to sialyl Lewis-x and glycopeptide conformation. There, I looked to see if single sugar could really force a random peptide to form a turn. The answer is a qualified "yes". Dr. Oliver Seitz welcomed me to the field.
I started my independent career at Texas A&M University where I remained on faculty for 12 years. I returned to the charge of "making something big," but instead of relying on transient, weak bonds between molecules, I chose polymer chemistry. The polymers, called dendrimers, are branched like trees. Motivated by a desire to impact human health, the metaphorical leaves on these trees were chosen to be anticancer drugs or bioactive groups. Collaborations around the world allowed us to explore the use of these polymers for many applications including fighting prostate cancer (with Drs. Xiankai Sun and JT Hsieh of UT Southwestern Medical Center & Anil Patri of NIH/FDA), for gene delivery (with Drs. Olivia Merkel and Thomas Kissel then of Marburg), and for fighting sepsis (with Dr. Sunil Shaunak at Imperial). "Curing" prostate cancer in mice with the group from UTSW was one of the highlights of these efforts.
The move to Texas Christian University allowed us to continue applied and basic inquiries into this system with a capstone accomplishment of the synthesis of one of (if not the) largest dendrimers to date, a molecule as big as a virus (30 nm across) with more than one million atoms.
The people who did it...